Gargle and mouthwash can replace nasopharyngeal swab sampling with a concentration method for diagnosis of COVID-19
Banu Sancak1, Esvet Mutlu2*, Osman Sezer Cirit3, Tanıl Kocagöz4, Özge Can5, Candan Çiçek6, Ayça Arzu Sayıner7, Özgür Appak7, Neval Yurttutan Uyar8, Canan Külah9, Ayşegül Çopur Çiçek10†, Osman Birol Özgümüş10, Yasemin Ay Altıntop11, Esma Saatçi11, Tekin Karslıgil12, Yasemin Zer12, Nevgün Sepin Özen13, Yeşim Çekin13, Zeynep Ceren Karahan14, Ebru Evren14, Ayşe Esra Karakoç15, Sultan Gülbahçe Orhan15, Derya Mutlu2, Tuğba Bozdemir2, Yeliz Tanrıverdi Çaycı16, Canberk Çınar16, Meltem Taşbakan17, Merve Mert17, Ece Çınar18, Oya Özlem Eren Kutsoylu19, Sesin Kocagöz20, Ayşe Ertürk21, İlhami Çelik22, Ayşe Özlem Mete23, Müge Günalp Eneyli24, İrem Akdemir25, Taliha Karakök26, Dilara İnan27, Aynur Atilla28
1 Hacettepe University, Faculty of Medicine Department of Medical Microbiology, Ankara, Türkiye
2 Akdeniz University, Faculty of Medicine Department of Medical Microbiology, Antalya, Türkiye
3 Gaziantep Dr. Ersin Arslan Training and Research Hospital, Medical Microbiology Laboratory, Gaziantep, Türkiye
4 Acibadem University, School of Medicine Department of Medical Microbiology and Medical Biotechnology, İstanbul, Türkiye
5 Acibadem University, School of Medical Engineering, İstanbul, Türkiye
6 Ege University, Faculty of Medicine Department of Medical Microbiology, İzmir, Türkiye
7 Dokuz Eylul University, Faculty of Medicine Department of Medical Microbiology, İzmir, Türkiye
8 Acibadem University, School of Medicine Department of Medical Microbiology, İstanbul, Türkiye
9 Acibadem Labmed, İstanbul, Türkiye
10 Recep Tayyip Erdogan University, Faculty of Medicine Department of Medical Microbiology, Rize, Türkiye
11 Kayseri City Training and Research Hospital, Medical Microbiology Laboratory, Kayseri, Türkiye
12 Gaziantep University, Faculty of Medicine Department of Medical Microbiology, Gaziantep, Türkiye
13 University of Health Sciences Antalya Training and Research Hospital, Medical Microbiology Laboratory, Antalya, Türkiye
14 Ankara University, Faculty of Medicine Department of Medical Microbiology, Ankara, Türkiye
15 University of Health Sciences Ankara Training and Research Hospital, Medical Microbiology Laboratory, Ankara, Türkiye
16 Ondokuz Mayis University, Faculty of Medicine Department of Medical Microbiology, Samsun, Türkiye
17 Ege University, Faculty of Medicine Department of Infectious Diseases and Clinical Microbiology, İzmir, Türkiye
18 Ege University, Faculty of Medicine Department of Physical Medicine and Rehabilitation, İzmir, Türkiye
19 Dokuz Eylul University, Faculty of Medicine Department of Infectious Diseases and Clinical Microbiology, İzmir, Türkiye
20 Acibadem University, School of Medicine Department of Infectious Diseases and Clinical Microbiology, İstanbul, Türkiye
21 Recep Tayyip Erdogan University, Faculty of Medicine Department of Infectious Diseases and Clinical Microbiology, Rize, Türkiye
22 Kayseri City Training and Research Hospital, Department of Infectious Diseases and Clinical Microbiology, Kayseri, Türkiye
23 Gaziantep University, Faculty of Medicine Department of Infectious Diseases and Clinical Microbiology, Gaziantep, Türkiye
24 Ankara University, Faculty of Medicine Department of Emergency Medicine, Ankara, Türkiye
25 Ankara University, Faculty of Medicine Department of Infectious Diseases and Clinical Microbiology, Ankara, Türkiye
26 University of Health Sciences Ankara Training and Research Hospital, Department of Infectious Diseases and Clinical Microbiology, Ankara, Türkiye
27 Akdeniz University, Faculty of Medicine Department of Infectious Diseases and Clinical Microbiology, Antalya, Türkiye
28 Ondokuz Mayis University, Faculty of Medicine Department of Infectious Diseases and Clinical Microbiology, Samsun, Türkiye
†Present address: İstanbul Medipol University, Faculty of Medicine, Department of Medical Microbiology, İstanbul, Türkiye
*Corresponding:
esvetmutlu[at]akdeniz.edu.tr ; dresvetmutlu[at]yahoo.com
ORCID: 0000-0001-8808-9182
Received: 06th March 2025
Accepted: 20th March 2025
Published: 08th April 2025
Abstract
Diagnosis of COVID19 is based on RT-PCR testing of nasopharyngeal swab samples. There is limited data about the performance of saliva and mouthwash samples by RT-PCR. We developed a new method for concentrating gargle and mouthwash samples to be used in RT-PCR. In our study, we aimed to investigate the performance of concentration of gargle and mouthwash samples in detection of SARS-CoV-2. Samples were collected from patients in 11 centers. Gargle and mouthwash samples were concentrated using MyMagiCon-RW100®. Nasopharyngeal swab, gargle and mouthwash and concentrated gargle and mouthwash samples were tested by RT-PCR for the presence of SARS-CoV2 and the results were compared. The viral RNA was detected in 47.5% of nasopharyngeal swab samples, in 28.8% of gargle and mouthwash before concentration and in 37.5% gargle and mouthwash samples after concentration. By concentration the number of SARS-CoV2 RNA positive samples increased 16.6%. Concentrated gargle and mouthwash sampling can be an alternative method to nasopharyngeal swab sampling in rapid and accurate diagnosis of COVID-19.
Keywords: Covid19, microorganism concentration, mouthwash, PCR, SARS-CoV-2
Abbreviations: COVID-19, coronavirus disease 2019; GMW, gargle and mouthwash; MMC-MW, mouthwash sample concentrated with MyMagiCon®; MW, mouthwash; NPS, nasopharyngeal swab; ONP, oronasopharyngeal; RT-PCR, reverse transcription-polymerase chain reaction; SARS-CoV-2, severe acute respiratory coronavirus 2.
Introduction
The pandemic of the coronavirus disease 2019 (COVID-19) which is caused by a severe acute respiratory coronavirus 2 (SARS-CoV-2) continues to infect many people worldwide. Currently, the diagnosis of COVID-19 is mainly based on reverse transcription-polymerase chain reaction (RT-PCR) testing.[1-2] The major clinical specimen for RT-PCR is nasopharyngeal swab (NPS) sampling which is an invasive method with complications like epistaxis. It requires expertized personnel and poses increased risk of viral transmission to the sampling person. Besides it requires continuous supplies of swabs and transport media. Therefore, alternative sample collection strategies which has less threat of transmission and more comfort are needed.
For detection of SARS-CoV-2 by PCR, which clinical specimen is more appropriate, is still unclear. Saliva or MW has many advantages over NPS because of easier collection of samples, having no need for experienced personnel and minimal requirement of supplies. Therefore, in various studies as a clinical sample for RT-PCR, saliva or mouth washing (MW) were investigated whether it is a reliable tool or not for COVID-19 diagnosis and monitoring when compared with gold standard samples (swab of nasopharynx or throat). Although there are numerous studies with saliva, only limited studies were found in literature with MW.[3-4]
In a previous study, we have tested the performance of a new method and tool, for concentrating liquid biological samples, which enables to use gargle and mouth-wash (GMW) samples in RT-PCR, as an alternative to NPS samples, which can be collected without the need of a health-personnel, in a limited number of patients.[5] MyMagiCon (Bio-T, Istanbul, Türkiye) is a biological fluid concentrator. It contains elastic polymer beads which absorb water and other small molecules with molecular weights less than 0.5kDa. Viruses and if disintegrated their nucleic acids and antigens are concentrated outside the beads. In this multicenter study, we aimed to investigate the efficiency of this method and product, in large number of patients, to assess its performance in different centers.
Materials and Methods
- Patients
The study was done between September 2020 and March 2021. A paired sample of GMW and NPS were prospectively collected from 11 centers in Türkiye. The patients presenting with signs/symptoms suggesting SARS-CoV-2 infection underwent a NPS and GMW collection. All the patients enrolled to the study, acknowledged understanding the aims of the study and signed a consent form prior to collection of clinical samples.
- Methods
After collecting NPS samples, patients were instructed to take a few sips of regular drinking water, and then to gargle and rigorously rinse their mouth forcefully with this water for at least 10 seconds and put it back to an empty sterile collection cup. 20 mL MW was collected.
Gargle and mouthwash samples were concentrated using MyMagiCon-RW100® (Bio-T, İstanbul, Türkiye) as instructed in the user guide. Briefly, 20 mL of sample was put into the tube and waited for 5 minutes for the absorbent beads to swell and absorb most of the fluid and the mixture turned in a gel-like form. Concentrated sample was collected with an automatic pipette by inserting the pipette tip in between the beads and aspirating the fluid (Figure 1). This concentrated sample was directly used in RT-PCR without RNA extraction.

Figure 1. Concentration method of gargle and mouthwash with MyMagicon RW100.
The RT-PCR was performed by using commercial PCR kits (Bioeksen and A1 Lifesciences, İstanbul, Türkiye) and the results obtained from NPS and GMW samples, before and after concentration, were compared. These kits included internal controls to detect the presence of inhibitors for amplification.
Since we have observed variability in the sensitivity of detection of SARS-CoV2 by RT-PCR in concentrated GMW between study centers we have also investigated the effect of the brand of drinking water used in different centers. For this purpose, we have prepared simulated GMW samples by adding SARS-CoV2 grown in cell culture, to GMW of healthy volunteers at the same concentration and studied them by RT-PCR and compared the Ct values.
- Statistical analysis
Categorical data were given in numbers and percentages and continuous variables were presented as median. Pearson Chi-square test and McNemar’s test were used for the diagnostic performance of PCR tests applied to NPS, MW and mouthwash sample concentrated with MyMagiCon® (MMC-MW), a p value less than or equal to 0.05 was considered significant. Statistical analyses were performed with a software (IBM SPSS Statistics for Windows, version 23.0).
Results
A total of 1721 patients were recruited into the study and samples pairs of NPS and GMW were collected. The PCR results of NPS, GMW and concentrated GMW of 11 centers are given in Table 1 and Figure 2.
Table 1. Sensitivity of COVID-19 PCR in NPS, GMW and Concentrated GMW Samples of each center (NPS: Nasopharyngeal swab; GMW: Gargle and mouthwash).
| Center | NPS | GMW | Concentrated GMW | Increase in sensitivity by concentration |
| A | 55(78.6%) | 30(42.9%) | 57 (81.4%) | 38.5% |
| B | 76 (66.7%) | 67 (58.8%) | 101 (88.6%) | 29.8% |
| C | 40 (85.1%) | 17 (36.2%) | 27 (57.4%) | 21.2% |
| D | 40 (88.9%) | 16 (35.6%) | 25 (55.6%) | 20.0% |
| E | 41 (100%) | 24 (58.5%) | 30 (73.2%) | 14.7% |
| F | 60 (93.8%) | 36 (56.3%) | 45 (70.3%) | 14% |
| G | 142 (95.3%) | 77 (51.7%) | 97 (65.1%) | 13.4% |
| H | 60 (98.4%) | 26 (42,6%) | 35 (57.4%) | 13.1% |
| I | 38 (88.4%) | 24 (55.8%) | 28 (65.1%) | 9.3% |
| J | 43 (100%) | 17 (39.5%) | 21 (48.8%) | 9.3% |
| K | 222 (99.1%) | 161 (71.9%) | 179 (79.9%) | 8% |
| Total | 817 (90.7%) | 495 (55%) | 645 (71.6%) | 16.6% |
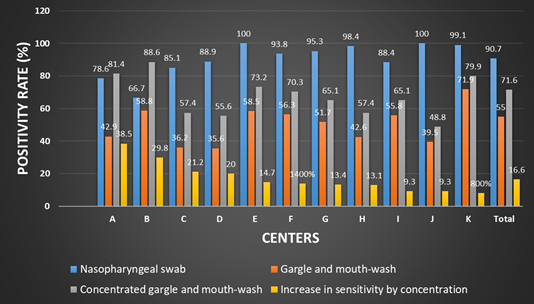
Figure 2. Sensitivity of COVID-19 PCR in nasopharyngeal, mouthwash, and concentrated mouthwash samples.
In all centers concentration of GMW samples increased the sensitivity of COVID-19 detection. It was determined that the PCR positivity between GMW and concentrated GMW, increased in 16.6 % (ranging from 8 to 38.5 % in different centers). In two centers concentrated GMW detected more positive cases than NPS samples.
When the results of 11 centers were evaluated, the percentage of PCR positivity of NPS, GMW and concentrated GMW showed prominent variability. This variability turned out to be mainly to the brand of water used in obtaining GMW samples.[5] Additionally due to the inclusion of only patients with positive NPS PCR in some centers.
The viral RNA was detected in 817 (90.7%) nasopharyngeal, in 495 (55%) gargle and mouthwash before concentration and in 645 (71.6%) after concentration among the total RT-PCR positive patients.
A total of 902 patients (52.4%) tested positive for the SARS-CoV-2 virus by NPS and/or GMW and/or concentrated GMW. In 43.3% patients SARS-CoV2 RNA was positive only in NPS samples. On the other hand it was positive in 3.4% and 9.0% of the GMW and concentrated GMW samples respectively, without being positive in NPS samples.
There was a moderate agreement between PCR tests using NPS and MMC-MW and significant difference in the detection rate (McNemar’s test κ=0.605, p<0.01). Concentration of samples increased the number of samples in which SARS-CoV-2 RNA was detected by 16.6 %. This increase varied between 8 and 38.5% among centers.
Discussion
Nasopharyngeal or throat swab are uncomfortable invasive procedures, which increase the infection risk to health care workers. Besides being time-consuming, it is labor intensive and depends on the availability of supplies such as nasal swabs and transport media. Therefore, an alternative diagnostic specimen which is noninvasive, easy to collect, cost-effective and having low COVID-19 infection risk for healthcare workers, is urgent necessity. For this purpose, saliva and mouthwash samples were used in different studies.[3-4,6,9-10] Although there are numerous studies with saliva, only limited studies were found with mouthwash samples. [3-4]
In our study we investigated the performance of MyMagiCon-RW100®, a new methodwhich enables concentrating GMW samples for increasing the sensitivity of detecting microorganism by RT-PCR. We also compared the diagnostic utility of GMW and concentrated GMW samples with gold standard sample, NPS.
Overall, concentrated GMW-based SARS-CoV-2 testing showed moderate concordance and sensitivity with nasal swab samples. However, the PCR of concentrated GMW samples yielded better results than the PCR of NPS in two centers. In all centers concentration of GMW samples increased the sensitivity of COVID-19 detection. The PCR positivity between GMW and concentrated GMW increased in 16.6 % (ranging from 8 to 38.5% in different centers).
In our previous study done in a single center the performance of the method differed from the total performance of the method done in this multicenter study.[5] When the results of each center examined separately, the percentage of PCR positivity of NPS, GMW and concentrated GMW showed variability. There can be several reasons for this variability. First of all, in some centers only patients with positive NPS were included. However, there can be COVID-19 patients with negative PCR results in NPS samples which was observed in centers which included all NPS samples either positive or negative by PCR. Secondly, we have
shown that different brands of drinking water used across the centers, which affected the RT-PCR amplification, is another explanation for false negative results. In some samples, even though the internal control showed amplification, partial inhibition of PCR may have led to false negative results, in samples containing low copy number of the virus. Therefore, adding a standard drinking water, which is shown not to inhibit PCR, as a part of MyMagiCon-RW100® kit can eliminate false negative PCR results. It will be beneficial, standardization of the collection of GMW sampling technique and the mouthwash fluid, in future studies.
Biber et al., also investigated the utility of using mouthwash (MW) samples for the detection of SARS-CoV2.[3] The patients either asymptomatic or with mild symptoms were included in the study. Although 84.6% (116/137) of oronasopharyngeal (ONP) swabs were positive by at least one of the genes (N, E, R), MW detected 70.8% (97/137) positivity. Unfortunately, MW samples missed samples especially NPS samples with Cq value >30. The median of Cq values of all positive ONP swabs (all three genes) was found as 30 compared to a median of 32 of positive MW. They also compared the results of different fluids (saline, distilled water, commercial mouthwash containing alcohol and commercial mouthwash without alcohol) for mouth rinsing.The result of water was similar to saline in contrast to other solutions, which had lower detection rate.
There are several studies which compared saliva to NPS and in most of them saliva sensitivity was found either comparable to that of NPS or higher than NPS. [11-20] Although there are several studies which compared saliva with NPS, only symptomatic patients were included in most of them. There are limited studies investigating the performance of saliva compared to NPS in asymptomatic individuals. [20-23] Alkhateeb et al., compared saliva and NPS of 33 symptomatic and 12 asymptomatic known SARS-CoV-2-positive patients.[21] Though, saliva showed lower sensitivity (36%) in asymptomatic patients than symptomatic patients (80%), it detected infections with lower Ct values. Opposite to the results of A1, there are researches in which saliva has same or sometimes higher sensitivity for detecting asymptomatic carriers compared to NPS.[21-24] Savela et al., evaluated the sensitivity and sample type for detecting early infections of COVID-19 in asymptomatic individuals.[24] They quantified SARS-CoV-2 RNA viral loads in anterior-nares nasal swabs and saliva samples which were obtained twice-daily and found that SARS-CoV-2 RNA first appears in saliva compared to nasal-swab samples. In other studies it has been demonstrated that SARS-CoV-2 RNA can appear in saliva 24–48 h prior to detection by nasopharyngeal swabs and 1.5–4.5 days prior to detection by anterior nasal swabs.[20,24-26] In the study of Yokoda et al., high sensitivity and specificity was detected in NPS and saliva specimens of 1924 asymptomatic individuals.[23]
One of the limitation of our study is including only symptomatic patients into the study. Overall, it is estimated that 17 to 30% of SARS-CoV-2 positive individuals remain asymptomatic.[26-27] As SARS-CoV-2 can spread from individuals before symptom onset and from asymptomatic individuals, the performance of GMW sampling in asymptomatic population must be also investigated.
Conclusion
The GWM sampling is a simple, rapid, cheap and self-collection method. According to the results of this study, concentrated GMW can be an alternative
sampling method to NPS in rapid and accurate diagnosis of COVID-19. Standardization of the method by using water with low ion concentration will increase the performance of the test and lower the variability of the results. Additional studies are needed including both symptomatic and asymptomatic patients for accurate evaluation of GMW and concentrated GMW sampling as a screening test for COVID-19.
Acknowledges
This study is organized by KLIMUD (Society for Clinical Microbiologists of Türkiye) research group.
MyMagiCon Kits were provided from Bio-T Biotechnology Solutions and RT-PCR kits were provided from Bioeksen and A1 Lifesciences.
Author Contributions
Banu Sancak: Writing of the first draft, methodology, conceptualization
Esvet Mutlu: Formal analysis, contribution to the final version of manuscript, data curation
Osman Sezer Cirit: Contribution to the final version of manuscript, data curation, visualization
Tanil Kocagöz: Project administration, supervision, writing, reviewing, and editing of the manuscript, methodology
Özge Can: Project administration
Candan Çicek: Investigation
Ayca Arzu Sayiner: Investigation
Özgür Appak: Investigation
Neval Yurttutan Uyar: Investigation
Canan Külah: Investigation
Aysegül Çopur Çiçek: Investigation
Osman Birol Özgümüs: Investigation
Yasemin Ay Altintop: Investigation
Esma Saatçi: Investigation
Tekin Karsligil: Investigation
Yasemin Zer: Investigation
Nevgün Sepin Özen: Investigation
Yesim Çekin: Investigation
Zeynep Ceren Karahan: Investigation
Ebru Evren: Investigation
Ayse Esra Karakoç: Investigation
Sultan Gülbahçe Orhan: Investigation
Derya Mutlu: Investigation
Tugba Bozdemir: Investigation
Yeliz Tanriverdi Çayci: Investigation
Canberk Çinar: Investigation
Meltem Tasbakan: Selection of patients, sample collection and transport to the laboratory
Merve Mert: Selection of patients, sample collection and transport to the laboratory
Ece Çinar: Selection of patients, sample collection and transport to the laboratory
Oya Özlem Eren Kutsoylu: Selection of patients, sample collection and transport to the laboratory
Sesin Kocagöz: Selection of patients, sample collection and transport to the laboratory
Ayse Ertürk: Selection of patients, sample collection and transport to the laboratory
Ilhami Çelik: Selection of patients, sample collection and transport to the laboratory
Ayse Özlem Mete: Selection of patients, sample collection and transport to the laboratory
Müge Günalp Eneyli: Selection of patients, sample collection and transport to the laboratory
Irem Akdemir: Selection of patients, sample collection and transport to the laboratory
Taliha Karakök: Selection of patients, sample collection and transport to the laboratory
Dilara Inan: Selection of patients, sample collection and transport to the laboratory
Aynur Atilla: Selection of patients, sample collection and transport to the laboratory
All authors have read and approved the final version of the manuscript.
Data Availability Statement
This study’s datasets are not publicly available, but can be provided by the Corresponding Author under a reasonable request.
Disclosure Statement
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflict of Interest
The authors Tanıl Kocagöz and Özge Can are scientific advisors of Bio-T.
Tanıl Kocagöz was a member of the Editorial Board of Bio&BioTech Journal at the time this article was written.
The other authors declare that they have no conflicts of interest.
Ethics Committee Approval
This study has been approved by the Acibadem University Ethics Committee, ATADEK-2020/14/2.
References
- Centers for Disease Control and Prevention (CDC). Interim Guidelines for Collecting, Handling, and Testing Clinical Specimens from Persons for Coronavirus Disease (2019) (COVID- 19), 2020. Last accessed date: 2020 April 14. Available from: https://www.cdc.gov/coronavirus/2019-nCoV/lab/guidelines-clinical-specimens.html.
- World Health Organization (WHO). Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases: interim guidance, 2020. Last accessed date: 2020 March 2. Available from: https://apps.who.int/iris/handle/10665/331329.
- Biber A, Lev D, Mandelboim M, Lustig Y, Harmelin G, Shaham A, Erster O, Schwartz E. Eur. J. Clin. Microbiol. Infect. Dis. 2021;40:2199-2206. https://doi: 10.1007/s10096-021-04320-4.
- Lai CK, Lui GC, Chen Z, Cheung YY, Cheng KC, Leung AS, Ng RW, Cheung JL, Yeung AC, Ho WC, Chan KC, Hui DS, Tsang DN, Chan PK. J. Infect. 2021;83:496-522. https://doi: 10.1016/j.jinf.2021.07.012.
- Kocagöz T, Can O, Yurttutan Uyar N, Aksoy E, Polat T, Cankaya D, Karakus B, Mozioglu E, Kocagoz S. Eur. Clin. Microbiol. Infect. Dis. 2021;40(12):2612-22. https://doi: 10.1007/s10096-021-04326-y.
- Caulley L, Shaw J, Corsten M, Hua N, Angel JB, Poliquin G, Whelan J, Antonation K, Johnson-Obaseki S. BMC Infect. Dis. 2021;21:410. https://doi: 10.1186/s12879-021-06108-5.
- Fernandes LL, Pacheco VB, Borges L, Athwal HK, de Paula Eduardo F, Bezinelli L, Correa L, Jimenez M, Dame-Teixeira N, Lombaert IMA, Heller D. J. Dent. Res. 2020;99:1435-43. https://doi: 10.1177/0022034520960070.
- Fernandes P, Ferreira F, Morais OM, Ramos CMT, Fernandes ÉMR, Rocha SAAD, Rocha RJA, Monteiro VJP, Vilar PSG, Romão AM, Alves MRA. J. Clin. Virol. 2021;142:104913. https:// doi: 10.1016/j.jcv.2021.104913.
- Plantamura J, Bousquet A, Otto MP, Bigaillon C, Legland AM, Delacour H, Vest P, Astier H, Valero E, Bylicki O, Renard C, Martin S, Verret C, Garnotel E, Foissaud V, Mérens A, Janvier F. Eur. J. Clin. Microbiol. Infect. Dis. 2021;40:2191-98. https:// doi: 10.1007/s10096-021-04269-4.
- Michel W, Farber J, Dilas M, Heuft HG, Tammer I, Baar J, Kaasch AJ. Infection. 2021:49:527-31. https://doi: 10.1007/s15010-021-01600-1.
- Abasiyanik MF, Flood B, Lin J, Ozcan S, Rouhani SJ, Pyzer A, Trujillo J, Zhen C, Wu P, Jumic S, Wang A, Gajewski TF, Wang P, Hartley M, Ameti B, Niemiec R, Fernando M, Mishra V, Savage P, Aydogan B, Bethel C, Matushek S, Beavis KG, Agrawal N, Segal J, Tay S, Izumchenko E. Sci. Rep. 2021;11:12425. https://doi: 10.1038/s41598-021-91835-7.
- Azzi L, Carcano G, Gianfagna F, Grossi P, Gasperina DD, Genoni A, Fasano M, Sessa F, Tettamanti L, Carinci F, Maurino V, Rossi A, Tagliabue A, Baj A. J. Infect. 2020; 81(1):e45-e50. https://doi: 10.1016/j.jinf.2020.04.005.
- Caulley L, Corsten M, Eapen L, Whelan J, Angel JB, Antonation K, Bastien N, Poliquin G, Johnson-Obaseki S. Ann. Intern. Med. 2021;174:131-3. https:// doi: 10.7326/M20-4738.
- Hanson KE, Barker AP, Hillyard DR, Gilmore N, Barrett JW, Orlandi RR, Shakir SM. J. Clin. Microbiol. 2020;58(11):e01824-20. doi: 10.1128/JCM.01824-20.
- Jamal AJ, Mozafarihashjin M, Coomes E, Powis J, Li AX, Paterson A, Anceva-Sami S, Barati S, Crowl G, Faheem A, Farooqi L, Khan S, Prost K, Poutanen S, Taylor M, Yip L, Zhong XZ, McGeer AJ, Mubareka S; Toronto Invasive Bacterial Diseases Network COVID-19 Investigators. Clin. Infect. Dis. 2021;72:1064-66. https://doi: 10.1093/cid/ciaa848.
- Sakanashi D, Asai N, Nakamura A, Miyazaki N, Kawamoto Y, Ohno T, Yamada A, Koita I, Suematsu H, Hagihara M, Shiota A, Kurumiya A, Sakata M, Kato S, Muramatsu Y, Koizumi Y, Kishino T, Ohashi W, Yamagishi Y, Mikamo H. J Infect Chemother. 2021;27:126-9. https:// doi: 10.1016/j.jiac.2020.09.027.
- Vaz SN, Santana DS, Netto EM, Pedroso C, Wang WK, Santos FDA, Brites C. Braz. J. Infect. Dis. 2020;24:422-7. https:// doi: 10.1016/j.bjid.2020.08.001.
- Kojima N, Turner F, Slepnev V, Bacelar A, Deming L, Kodeboyina S, Klausner JD. Clin. Infect. Dis. 2021;Nov 2, 73(9): e3106-e3109.https://doi.org/10.1093/cid/ciaa1589.
- Moreno-Contreras J, Espinoza MA, Sandoval-Jaime C, Cantú-Cuevas MA, Barón-Olivares H, Ortiz-Orozco OD, Muñoz-Rangel AV, Hernández-de la Cruz M, Eroza-Osorio CM, Arias CF, López S. J. Clin. Microbiol. 2020;22:58(10):e01659-20. https://doi.org/10.1128/JCM.01659-20.
- Wyllie AL, Fournier J, Casanovas-Massana A, Campbell M, Tokuyama M, Vijayakumar P, Warren JL, Geng B, Muenker MC, Moore AJ, Vogels CBF, Petrone ME, Ott IM, Lu P, Venkataraman A, Lu-Culligan A, Klein J, Earnest R, Simonov M, Datta R, Handoko R, Naushad N, Sewanan LR, Valdez J, White EB, Lapidus S, Kalinich CC, Jiang X, Kim DJ, Kudo E, Linehan M, Mao T, Moriyama M, Oh JE, Park A, Silva J, Song E, Takahashi T, Taura M, Weizman OE, Wong P, Yang Y, Bermejo S, Odio CD, Omer SB, Dela Cruz CS, Farhadian S, Martinello RA, Iwasaki A, Grubaugh ND, Ko AI. N. Engl. J. Med. 2020;383:1283-86. https://doi:10.1056/NEJMc2016359.
- Alkhateeb KJ, Cahill MN, Ross AS, Arnold FW, Snyder JW. Diagn. Microbiol. Infect. Dis. 2021;101:115450. https:// doi: 10.1016/j.diagmicrobio.2021.115450.
- Vogels CBF, Watkins AE, Harden CA, Brackney DE, Shafer J, Wang J, Caraballo C, Kalinich CC, Ott IM, Fauver JR, Kudo E, Lu P, Venkataraman A, Tokuyama M, Moore AJ, Muenker MC, Casanovas-Massana A, Fournier J, Bermejo S, Campbell M, Datta R, Nelson A; Yale IMPACT Research Team, Dela Cruz CS, Ko AI, Iwasaki A, Krumholz HM, Matheus JD, Hui P, Liu C, Farhadian SF, Sikka R, Wyllie AL, Grubaugh ND. Med (N Y). 2021;2:263-80 e6 https://doi: 10.1016/j.medj.2020.12.010.
- Yokota I, Shane PY, Okada K, Unoki Y, Yang Y, Inao T, Sakamaki K, Iwasaki S, Hayasaka K, Sugita J, Nishida M, Fujisawa S, Teshima T. Clin. Infect. Dis. 2021;73:e559-e65. https:// doi: 10.1093/cid/ciaa1388.
- Savela ES, Winnett A, Romano AE, Porter MK, Shelby N, Akana R, Ji J, Cooper MM, Schlenker NW, Reyes JA, Carter AM, Barlow JT, Tognazzini C, Feaster M, Goh YY, Ismagilov RF. Quantitative SARS-CoV-2 viral load curves in paired saliva and nasal swabs inform appropriate respiratory sampling site and analytical test sensitivity required for earliest viral detection. medRxiv. 2021;7:2021.04.02.21254771. https:// doi: 10.1101/2021.04.02.21254771. Preprint.
- Borghi E, Massa V, Zuccotti G, Wyllie AL. Front. Microbiol. 2021;12:721635. https:// doi: 10.3389/fmicb.2021.721635.
- Pollock A. M., Lancester J. BMJ. 2020;37:4851 https://dx.doi.og/10.1136/bmj.m4851
- Johansson MA, Quandelacy TM, Kada S, Prasad PV, Steele M, Brooks JT, Slayton RB, Biggerstaff M, Butler JC. 2021;4:e2035057. https://doi:10.1001/jamanetworkopen.2020.35057.
